What is imputation?
Imputation covers a set of approaches for taking missing values in a matrix (e.g. a counts matrix of genes by cells) and filling them in. One of the most famous examples of an imputation problem is The Netflix Problem. Here, you have a matrix where the rows are users and the columns are movies they’ve liked watching. Let’s call this the user-ratings matrix. You can imagine that the user-ratings matrix starts out filled with zeros. After each user watches a movie, they click on a thumbs-up for thumbs-down button and changes a single value of the matrix to +1 (like) or -1 (dislike).
Because not all users watch all movies, you might imagine that after a year, the user-ratings matrix is 80% zeros and 20% +1 or -1. It might look like the following matrix:
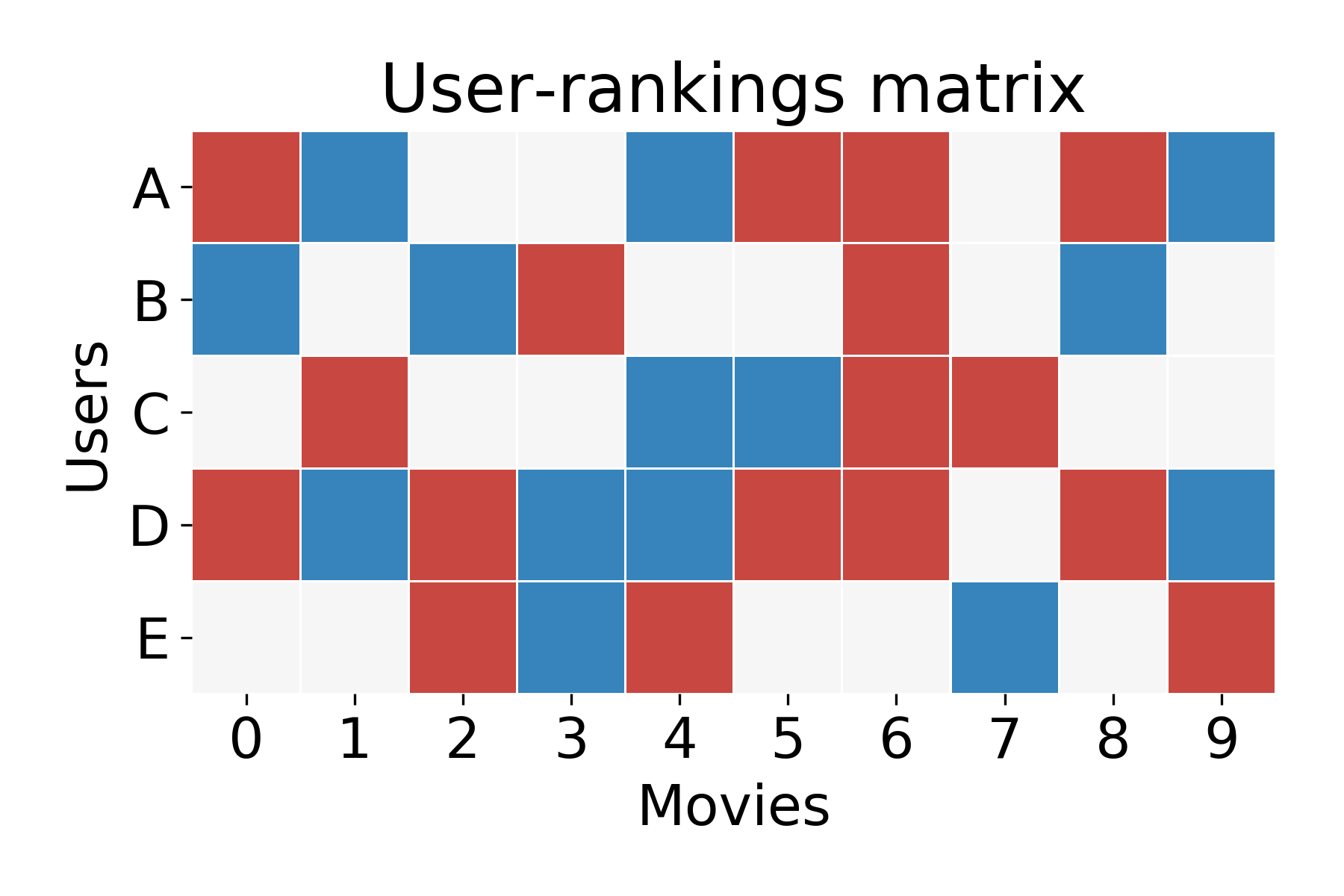
Now, the Netflix Problem asks, how can I guess if User A will like or dislike a movie, let’s call it Movie 1. they haven’t seen? Because User A hasn’t seen Movie 1, the corresponding cell of the user-ratings matrix will be zero. You can think of this as missing data. There is definitely a true value that exists in that cell (i.e. if User A watched Movie 1 they would either like or dislike it), but we don’t know what it is.
Can’t we just use average rankings?
A naive approach might be to just take the average ranking for each movie. Here, you simply take the average of the non-zero values for each column, and replace the zeros with that average. This naive approach might product a matrix like the following:
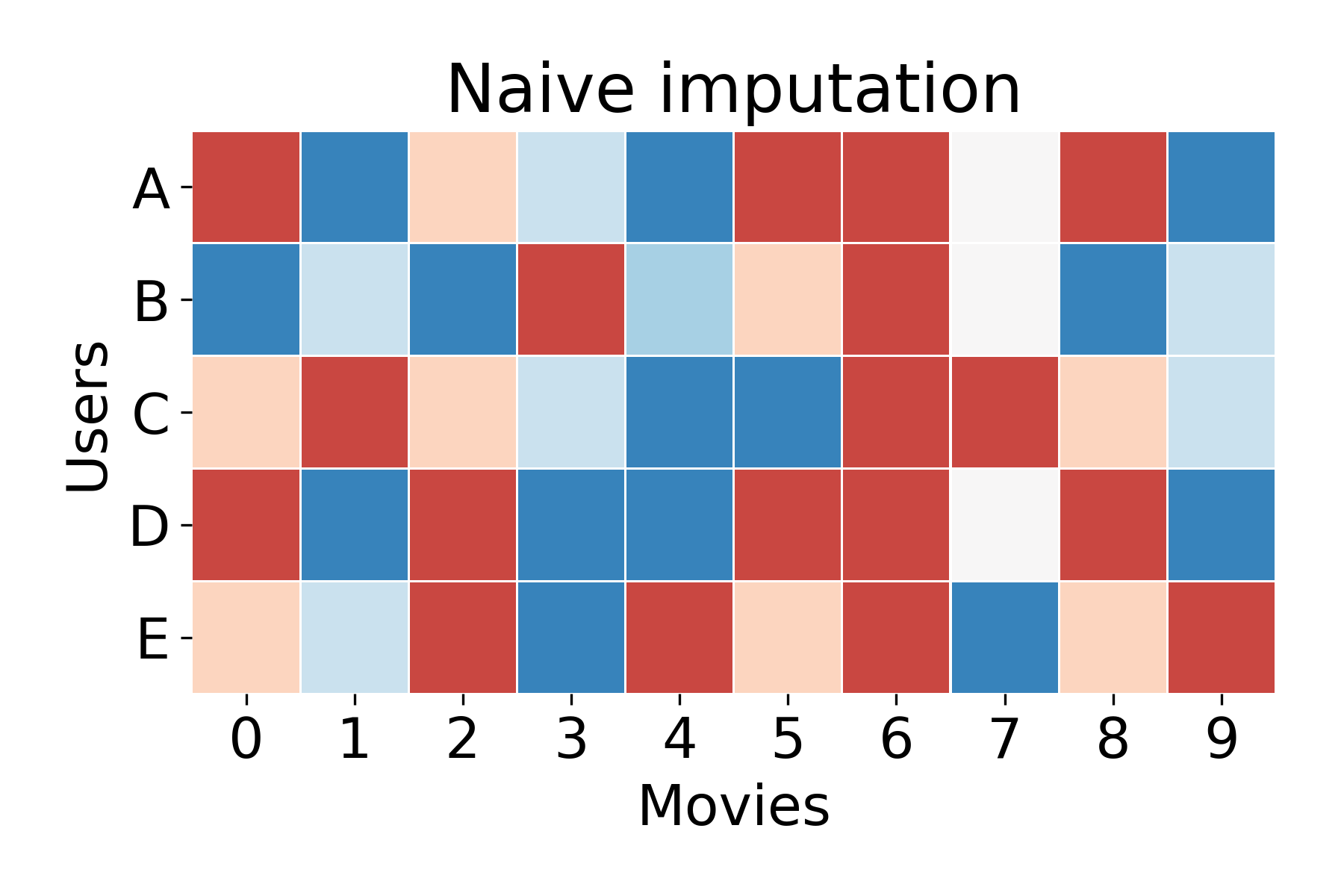
Although this is probably the best approach to take for a new user about whom we have no information, shouldn’t we be able to do better for a user who’s preferences we already know? Say, for example, we’re interested in User A who’s watched 7⁄10 movies in our library. We should be able to predict what User A will like more accurately than we can for a new user.
A more sophisticated approach
Intuitively, you might think, well, let’s just find a similar User Y who has:
1) watched a bunch of the same users as User A
2) rated those shared movies similarly to User A
In our raw user-rankings matrix, you might notice that User D satisfies these criteria well.
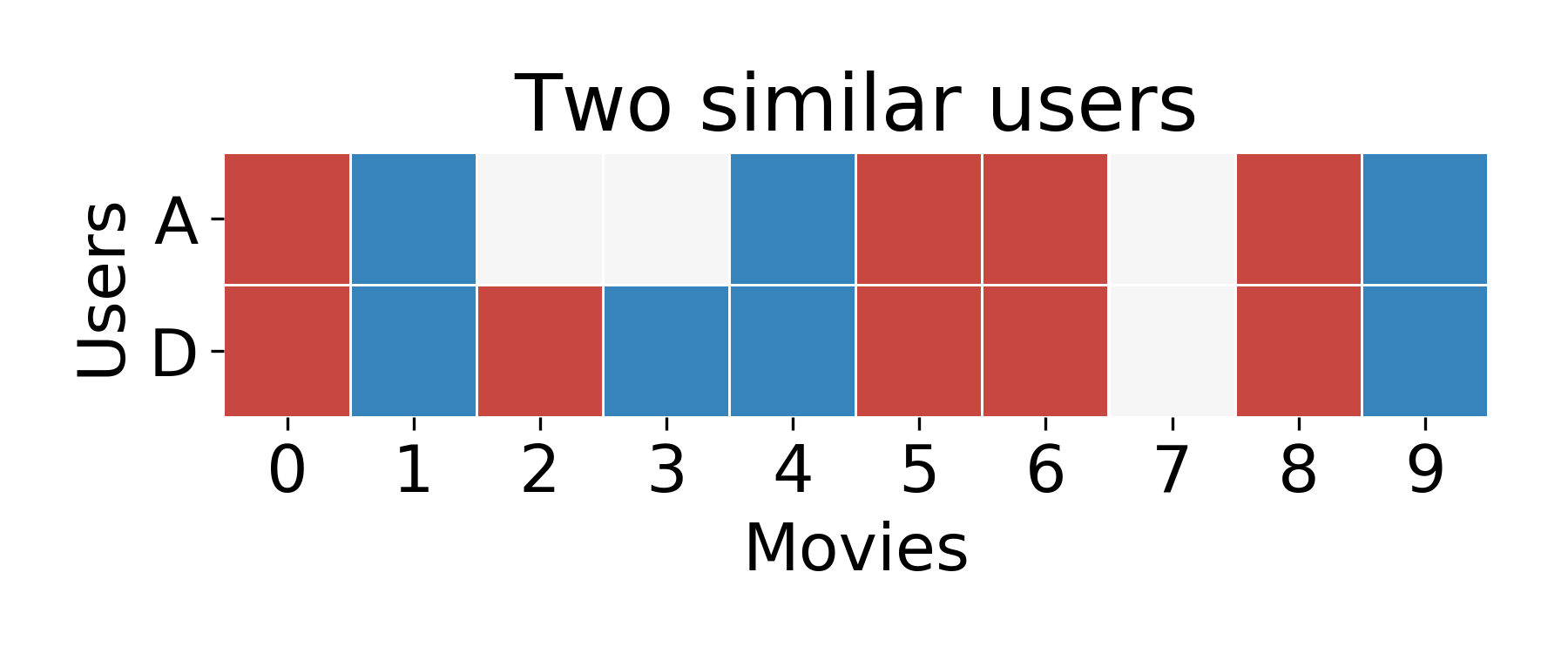
Now, all we have to do is guess that User A will like the same movies that User D did!
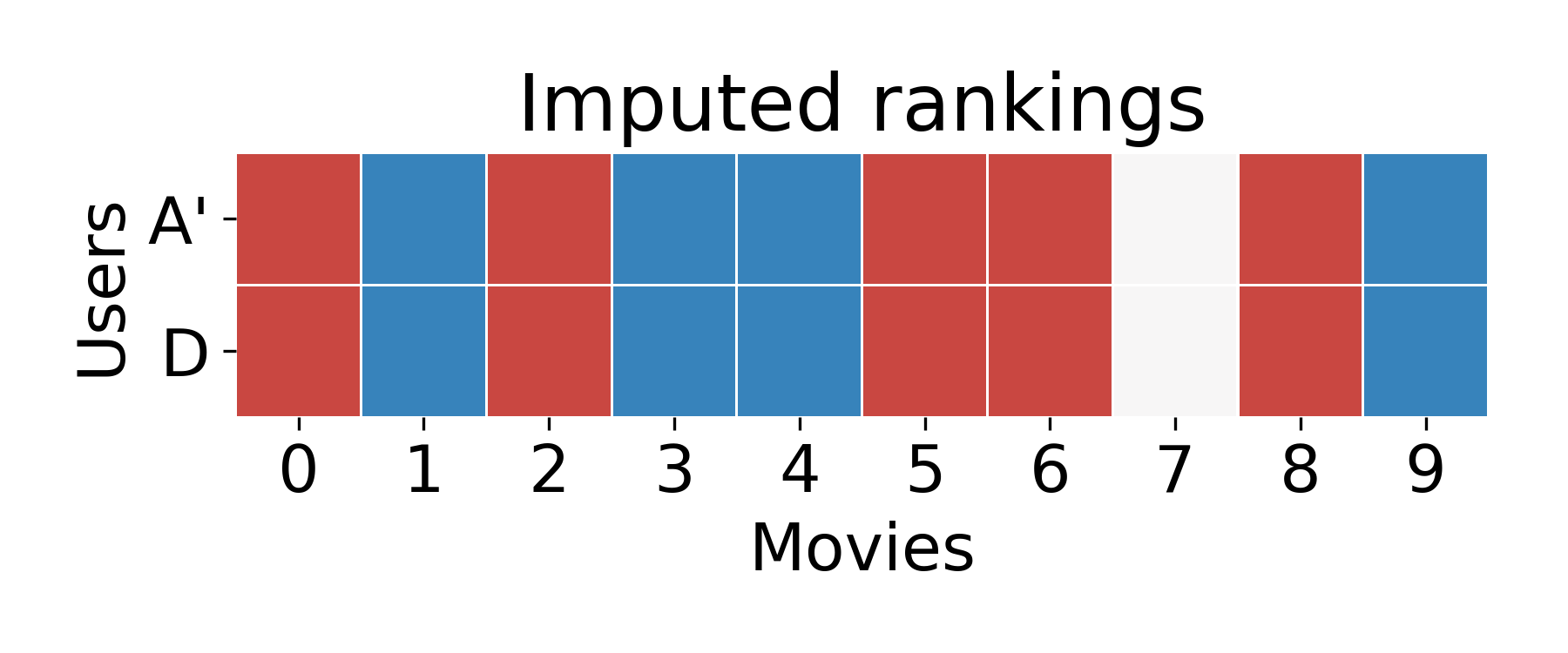
Matrix completion
Here, we’re taking advantage of the knowledge that two users are similar to infer what movies User A will like to watch. We might think of this as a form of redundancy among the users of the matrix. If we had more users, we might take the average of 10 users that are similar to User A and average their likes/dislikes. To go even further, perhaps we can take a weighted average where the preferences of users that are more similar to User A count more in coming up with a ranking. Surely this would perform better than just averaging over the whole population.
You may have already realized this, but there’s an equally important kind of redundancy in this matrix: similarities among the movies. For example, if a user likes rom-coms but hates horror movies, then we should probably be recommending more rom-coms to that user. Importantly, we can learn these relationships from the user-rankings matrix.
As described here, we can consider imputation as a form of matrix-completion. There are many ways to solve the matrix completion problem, and one elegant solution proposed by Candes and Recht (2008) considers the rank of the matrix. The rank of a matrix is equal to the number of linearly independent columns. In the Netflix Problem, we assume that the ratings of all rom-coms are not linearly independent, but are to some degree related to each other.
Single cell imputation is not a matrix completion
As you may have realized, single cell imputation is not a simple matrix completion problem. Can you tell why?
The fundamental issue is that we don’t know which values in the matrix are missing. In the Netflix Problem, we know which movies a user has and has not watched, but in scRNA-seq, we don’t know which genes are true zeros (i.e. not expressed) and which are missing data due to noise.
Because we don’t know which zeros to trust, we need a more sophisticated approach to scRNA-seq imputation. This being said, we can still rely on the sources of redundancy in the rows (cells) and columns (genes).
In the next part of our tutorial we will focus on an imputing gene expression in single cells. We’ll focus on a method called MAGIC (Markov Affinity Graph Imputation of Cells) developed by our lab van Dijk, D. et al. 2018.. We’re not going to do a full comparison of imputation methods here, so we encourage you to test out different algorithms yourself.